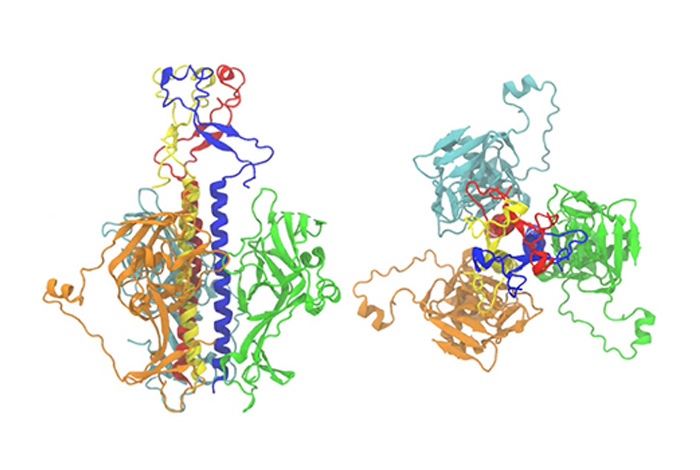
Researchers at Los Alamos National Laboratory (LANL) recently used computer modeling to map the process by which the Ebola and Zika viruses infiltrate host cells.
The researchers, who published their work in the journal Biomolecules, aimed to understand the specific structure-function relationship of the Ebola glycoprotein (EBOV GP) and Zika envelope (ZIKA E) proteins, which enable fusion with the host cell. This improved understanding could aid in the development of vaccines and therapeutic medicines.
The team used a computational approach developed at LANL by Chang-Shung Tung, the Motif-Matching Fragment Assembly (MMFA) method, to combine the available structural information from both low- and high-resolution experimental protein structures.
“Before the membrane fusion started, these two proteins went through large conformational changes that was not fully understood, as the proteins would take on the physical shape that would allow them to fuse with the cell and begin the infection,” Tung, the leading and corresponding author of the paper, said. “In this work, we modeled the structures of the two proteins in the fusion-initiation state. Then we used the structures to explain how four different antibodies in our bodies can fight the infection and block the viral entry, based on how they bind to these two proteins.”
The research team discovered that some of the antibodies prevent cell entry by blocking the “pre-fusion to fusion” transition, others prevent the exposure of the fusion peptide at the tip of the molecule when the envelope proteins are in the fusion state and others take on both roles.
“This computer modeling approach allowed us to generate atomic-level models of EBOV GP and ZIKV E proteins in the fusion state for the first time,” Anna Lappala, a postdoctoral fellow at the Center for Nonlinear Studies and the Theoretical Biology & Biophysics Group at Los Alamos, said. “These models, together with the knowledge of the envelope proteins in the pre-fusion state, allowed us to study the function of neutralizing antibodies as they bind to the viral envelope proteins.”
The modeling approach, Lappala said, will be useful for predicting and assessing antibody binding to the viral proteins, which could have vast medical applications.